Pfeifer
Laboratory
 Epigenetic and Genetic Pathways in Human Disease
Epigenetic and Genetic Pathways in Human Disease
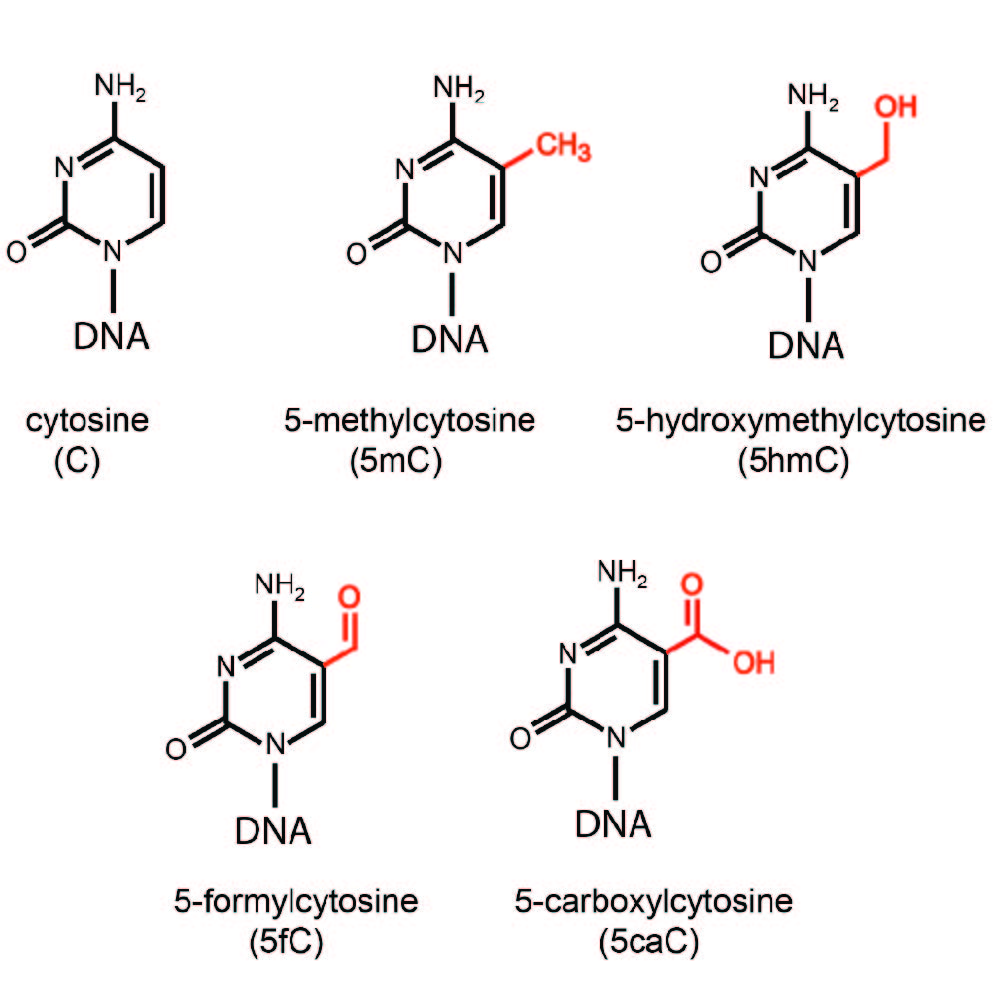
The Pfeifer Laboratory studies how epigenetic and genetic processes contribute to the etiology of cancer and potentially to other human diseases. One major research direction aims to understand the mechanisms of DNA methylation and demethylation, two antagonistic pathways that are disrupted in disease and lead to aberrant cell function and malignant transformation. DNA demethylation involves enzymatic oxidation of 5-methylcytosine (5mC) by TET dioxygenases leading to the formation of 5-hydroxymethylcytosine (5hmC) and additional oxidation products that are subsequently removed by DNA repair. The laboratory investigates how the function of TET proteins is controlled, how DNA methylation patterns are altered in disease and what consequences these alterations have for cellular phenotypes. In another research direction, the laboratory aims to develop plausible models and mechanisms of how specific mutational signatures arise in selected types of human cancer as a consequence of DNA damage.
Recent News & Press
Learn MoreOur Impact
We’re raising thousands to save millions.
We’re turning hope into action for the millions of people around the world affected by diseases like cancer and Parkinson’s. Find out how you can help us make a difference.
- 122 peer-reviewed papers published in 2024, 63 of which were in high-impact journals
- 15 VAI-SU2C Epigenetics Dream Team clinical trials launched to date
- 10 clinical trials co-funded by VAI & Cure Parkinson's (out of 41 total International Linked Clinical Trials Program trials)
Gerd Pfeifer, Ph.D.
Professor, Department of Epigenetics
Areas of Expertise
Epigenetics, DNA methylation, 5-hydroxymethylcytosine, mutagenesis
Biography
Dr. Gerd Pfeifer earned his Ph.D. in biochemistry from Goethe University in Frankfurt, Germany. Following a postdoctoral fellowship at the University of Frankfurt Medical School, he joined the Beckman Research Institute at City of Hope in Duarte, Calif. in 1988. In 1993, he became full member of City of Hope Comprehensive Cancer Center and, in 1999, he was promoted to Professor of Biology at Beckman Research Institute. He also served as Program Leader for the City of Hope’s Programs in DNA Damage and Repair, Cancer Biology, and Lung Cancer; as chair of the Department of Biology (2001–2008); and as chair of the Department of Cancer Biology (2008–2012). At City of Hope, he was the Lester M. and Irene C. Finkelstein Chair in Biology (2008–2014). He joined VAI in 2014 as a Professor in the Department of Epigenetics. In 2015, he was elected as Fellow of the American Association for the Advancement of Science (AAAS).
DNA methylation and cancer
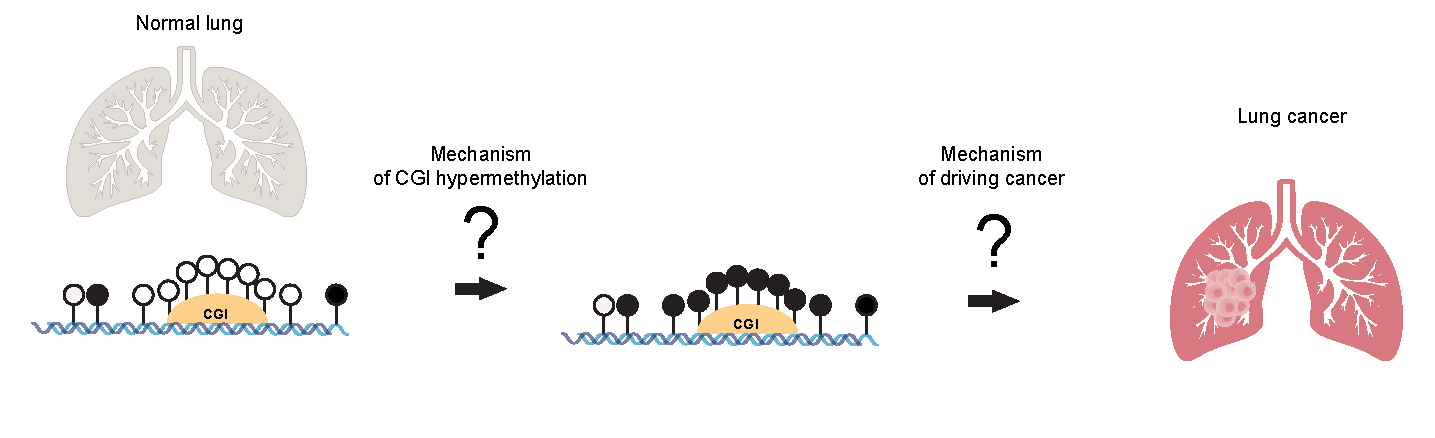
One of the lab’s primary research projects is studying the mechanisms of DNA hypermethylation in tumors and the biological significance of hypermethylation events in cancer by investigating the molecular pathways that lead to DNA methylation changes in tumors. The lab’s hypothesis is that specific factors such as carcinogenic agents, oncogene activation, inflammation, mechanisms related to the Polycomb repression complex, and imbalances between methylation and demethylation pathways drive the common event of DNA hypermethylation in cancer.
One study focuses on lung tumors. In lung cancers, as in most other cancer types, CpG-rich DNA regions (CpG islands) are frequently methylated in tumors. However, the mechanisms of this DNA hypermethylation isunknown. Since around a thousand or more genes become methylated in tumors, there is a need and a challenge to understand which ones of these many epigenetic events are truly cancer driving by altering the expression of genes critical for normal and malignant cell function.
The role of 5-methylcytosine oxidation in development and disease
The Pfeifer Lab has previously found that 5-hydroxymethylcytosine (5hmC) is strongly depleted in human cancer. The lab hypothesizes that defects in 5-methylcytosine (5mC) oxidation are responsible for different DNA methylation patterns in tumors and, potentially, other diseases.
To test this hypothesis, the Pfeifer Lab has established and used methodology for genome-wide mapping and precise quantification of 5mC and 5hmC, with the goal of determining genomic distribution of these modified DNA bases in normal tissues, in malignant tumors and in other disease states. To this end, the lab focuses on solid tumors and investigates the role of the TET proteins that catalyze the conversion of 5mC to 5hmC and additional oxidized bases. The lab conducts basic mechanistic studies of TET and TET-associated proteins, along with their regulation, abnormalities in cancer and their roles in cell differentiation and epigenome integrity. One study focuses on the chromosomal protein SMCHD1, which is mutated in a form of muscular dystrophy. We characterize the role of SMCHD1 in human muscle cells and in embryonic stem (ES) cells, where this protein regulates DNA methylation patterns, gene expression, and the two-cell-like state of ES cells (Huang et al., 2021).
In muscle cells, SMCHD1 plays a prominent role in maintaining the three-dimensional organization of chromosomes and serves as an anchor for silent regions of the genome (called heterochromatin) at the nuclear lamina, a structure that surrounds the cell nucleus in human cells (Huang et al., 2025).

Origins of mutations in cancer
Cancer genomes are characterized by large numbers of mutations with very different mutational changes (mutational signatures) found in different tumor types. With the exception of sunlight-induced skin cancers and smoking-associated lung cancers, there are only a few other cases where the mechanisms of the mutations’ origin are understood. The lab focuses on a few cancer types that include liver cancer and esophageal cancer, for which the mechanisms of mutation are unknown, but for which models can be proposed based on the mutation data available. Toward analyzing novel mechanisms of mutagenesis, the Pfeifer Lab has recently developed new, highly sensitive methodology to characterize specific DNA damage at every position in the human genome. This method called “circle-damage-sequencing” (or “CD-seq”) is applicable to many types of DNA damage that can be produced experimentally or may even exist at certain levels as endogenous DNA lesions (Jin et al., 2021, 2022).

UVA radiation and melanoma
Melanoma is a lethal type of skin tumor that has been linked with sunlight exposure chiefly in fair-skinned human populations. Wavelengths from the sun that can reach the earth’s surface include UVA radiation (320 to 400 nm) and UVB radiation (280 to 320 nm). UVB effectively induces the formation of dimeric DNA photoproducts, preferentially the cyclobutane pyrimidine dimers (CPDs). The characteristic UVB signature mutations in the form of C to T mutations at dipyrimidine sequences are readily apparent in melanoma tumor genomes and have been ascribed to deamination of cytosines within CPDs before DNA polymerase bypass (Jin et al., 2021). However, evidence from epidemiological, animal, and other experimental studies suggest that UVA radiation also participates in melanoma formation. The DNA damage relevant for UVA includes specific types of pyrimidine dimers at TT sequences and oxidative DNA damage to guanine, both induced by direct or indirect, photosensitization-mediated chemical and biophysical processes. We investigate the potential role of UVA in melanoma and try to understand the mechanistic pathways of how UVA may induce mutagenesis in melanocytes, with special emphasis on the role of the different types of melanin in these photocarcinogenic processes.

SELECTED PUBLICATIONS
For a full list of Dr. Pfeifer’s publications, please visit PubMed.
Huang Z, Cui W, Ratnayake I, Gallik KL, Cohen L, Tawil R, Pfeifer GP. 2025. SMCHD1 maintains heterochromatin, genome compartments and epigenome landscape in human myoblasts. Nat Commun 16:6900.
Pfeifer GP, Jin SG. 2024. Methods and applications of genome-wide profiling of DNA damage and rare mutations. Nature Rev Genet 25(12):846–863.
Blanchett R, Lau KH, Pfeifer GP. 2024. Homeobox and Polycomb target gene methylation in human solid tumors. Sci Rep 14(1):13912.
Pfeifer GP. 2024. DNA damage and Parkinson’s disease. Int J Mol Sci 25(8):4187.
Jin SG, Johnson J, Huang Z, Cui W, Dunwell T, Pfeifer GP. 2024. CXXC5 stabilizes DNA methylation patterns in mouse embryonic stem cells. Epigenomics 16(21–22):1351–1363.
Cui W, Huang Z, Jin S-G, Johnson J, Lau KH, Hostetter G, Pfeifer GP. 2023. Deficiency of the Polycomb protein RYBP and TET methylcytosine oxidases promotes extensive CpG island hypermethylation and malignant transformation. Cancer Res.
TRIM28 secures skeletal stem cell fate during skeletogenesis by silencing neural gene expression and repressing GREM1/AKT/mTOR signaling axis. Cell Rep 42(1):112012.
Jin SG, Padron F, Pfeifer GP. 2022. UVA radiation, DNA damage, and melanoma. ACS Omega 7(37):32936-32948.
Meng Y, Wang G, He H, Lau KH, Hurt A, Bixler BJ, Parham A, Jin SG, Xu X, Vasquez KM, Pfeifer GP, Szabó PE. 2022. Z-DNA is remodelled by ZBTB43 in prospermatagonia to safeguard the germline and epigenome. Nat Cell Biol.
Jin SG, Meng Y, Johnson J, Szabó PE, Pfeifer GP. 2022. Concordance of hydrogen peroxide-induced 8-oxoguanine patterns with two cancer mutation signatures of upper GI tract tumors. Sci Adv 8(2).
Pfeifer GP. 2022. The ups and downs of DNA methylation: an interview with Gerd Pfeifer. Epigenomics 14(6):339–343.
Cui W, Huang Z, Pfeifer GP. 2022. Lack of major genome-wide DNA methylation changes in succinate-treated human epithelial cells. Int J Mol Sci 23(10):5663.
Pfeifer GP. 2021. DNA repair in neurons and its possible link to the epigenetic machinery at enhancers. Epigenomics 13(12): 913–917.
Kim SI, Pfeifer GP. 2021. The epigenetic DNA modification 5-carboxylcytosine promotes high levels of cyclobutane pyrimidine dimer formation upon UVB irradiation. Gen Insta Dis 2(1):59–69.
Jin SG, Pettinga D, Johnson J, Li P, Pfeifer GP. 2021. The major mechanism of melanoma mutations is based on deamination of cytosine in pyrimidine dimers as determined by circle damage sequencing. Sci Adv 7(31):eabi6508.
Huang Z, Yu J, Johnson J, Jin SG, Pfeifer GP. 2021. Purification of TET proteins. Methods Mol Biol 2272:225–237.
Huang Z, Yu J, Cui W, Johnson BK, Kim K, Pfeifer GP. 2021. The chromosomal protein SMCHD1 regulates DNA methylation and the 2c-like state of embryonic stem cells by antagonizing TET proteins. Sci Adv 7(4): eabb9149.
Huang Z, Meng Y, Szabó PE, Kohli RM, Pfeifer GP. 2021. High-resolution analysis of 5-hydroxymethylcytosine by TET-assisted bisulfite sequencing. Methods Mol Biol 2198:321–331.
Himadewi P, Wang XQD, Feng F, Gore H, Liu Y, Yu L, Kurita R, Nakamura Y, Pfeifer GP, Zhang X. 2021. 3’HS1 CTCF binding site in human β-globin locus regulates fetal hemoglobin expression. eLife 10:e70557.
Pfeifer GP. 2020. Smoke signals in the DNA of normal lung cells. Nature 578:224–226.
Pfeifer GP. 2020. Mechanisms of UV-induced mutations and skin cancer. Genome Instab Dis 1:9–113.
Pfeifer GP, Szabó PE, Song J. 2020. Protein interactions at oxidized 5-methylcytosine bases. J Mol Biol 432(6):1718–1730.
Zeng TB, Han L, Pierce N, Pfeifer GP, Szabó PE. 2019. EHMT2 and SETDB1 protect the maternal pronucleus from 5mC oxidation. Proc Natl Acad Sci U S A 116:10834–10841.
Pfeifer GP, Szabó PE, Song J. 2020. Protein interactions at oxidized 5-methylcytosine bases. J Mol Biol 432(6):1718-1730.
Hahn MA, Jin SG, Li AX, Liu J, Huang Z, Wu X, Byung-Wook K, Johnson J, Bilbao ADV, Tao S, Yim JA, Fong Y, Goebbels S, Schwab MH, Lu Q, Pfeifer GP. 2019. Reprogramming of DNA methylation at NEUROD2-bound sequences during cortical neuron differentiation. Sci Adv 5(10):eaax0080.
Yim JH, Choi AH, Li AX, Qin H, Chang S, Tong ST, Chu P, Kim BW, Schmolze D, Lew R, Ibrahim Y, Poroyko VA, Salvatierra S, Baker A, Wang J, Wu X, Pfeifer GP, Fond Y, Hahn MA. 2019. Identification of tissue-specific DNA methylation signatures for thyroid nodule diagnostics. Clin Cancer Res 25(2):544–551.
Li W, Yue F, Dai Y, Shi B, Xu G, Jiang X, Zhou X, Pfeifer GP, Liu L. 2019. Suppressor of hepatocellular carcinoma RASSF1A activates autophagy initiation and maturation. Cell Death Diff 26(8):1379–1395.
Hahn MA, Jin SG, Li AX, Liu J, Huang Z, Wu X, Kim BW, Johnson J, Bilbao ADV, Tao S, Yim JA, Fong Y, Goebbels S, Schwab MH, Lu Q, Pfeifer GP. 2019. Reprogramming of DNA methylation at NEUROD2-bound sequences during cortical neuron differentiation. Sci Adv 5(10):eaax0080.
Pfeifer GP, Szabó PE. 2018. Gene body profiles of 5-hydroxymethylcytosine: potential origin, function and use as a cancer biomarker. Epigenomics.
Pfeifer GP. 2018. Defining driver DNA methylation changes in human cancer. Intl J Mol Sci 19(4):1166.
Liu J, Wu X, Zhang H, Pfeifer GP, Lu Q. 2017. Dynamics of RNA polymerase II pausing and bivalent histone H3 methylation during neuronal differentiation in brain development. Cell Rep 20(6)1307–1318.
Hanley MP, Hahn MA, Li AX, Wu X, Lin J, Wang J, Choi A, Ouyang Z, Fong Y, Pfeifer GP, Devers TJ, Rosenberg DW. 2017. Genome-wide DNA methylation profiling reveals cancer-associated changes within early colonic neoplasia. Oncogene 36(35):5035–5044.
Weng YL, An R, Cassin J, Joseph J, Mi R, Wang C, Zhong C, Jin SG, Pfeifer GP, Bellacosa A, Dong X, Hoke A, He Z, Song H, Ming GL. 2017. An intrinsic epigenetic barrier for functional axon regeneration. Neuron 94(2):337–346.
Pfeifer GP. 2017. DNA methylation and mutation 2.0. Encyclopedia of Life Sciences. John Wiley & Sons, Ltd, Chichester.
Rauch TA, Pfeifer GP. 2017. Methods for assessing DNA cytosine modifications genome-wide. Handbook of Epigenetics, 2nd Edition, edited by T. Tollefsbol, Elsevier. Waltham, MA. pp. 125–134.
Pfeifer GP, Kernstine KH. 2017. DNA methylation biomarkers in lung cancer diagnosis: closer to practical use? Transl Cancer Res 6(Suppl. 1):S122–S126.
Pfeifer GP. 2016. How tobacco smoke changes the (epi)genome. Science 354(6312):549–550.
Song J, Pfeifer GP. 2016. Are there specific readers of oxidized 5-methylcytosine bases? BioEssays 38(10):1038–1047.
Iqbal K, Tran DA, Li AX, Warden C, Bai AY, Singh P, Madaj ZB, Winn ME, Wu X, Pfeifer GP, Szabó PE. 2016. High type I error and misrepresentations in search for transgenerational epigenetic inheritance: response to Guerrero-Bosagna. Genome Biol 17:154.
Pfeifer GP. 2016. Switching enhancer methylation in metastatic melanoma. Pigm Cell Melanoma Res 29(5):491 –493.
Pfeifer GP. 2016. Epigenetics: An elusive DNA base in mammals. Nature 532:319–320.
Zhang X, Guo C, Wu X, Li AX, Liu L, Tsark W, Dammann R, Shen H, Vonderfecht SL, Pfeifer GP. 2016. Analysis of liver tumor-prone mouse models of the Hippo kinase scaffold proteins RASSF1A and SAV1. Cancer Res 76:2824–2835.
Pfeifer GP. 2016. Properly dividing with YAP. Sci Signal 9(417):fs3.
Hainaut P, Pfeifer GP. 2016. Somatic TP53 mutations in the era of genome sequencing. Cold Spring Harb Perspect Med 6(11).
Jin SG, Zhang ZM, Dunwell TL, Harter MR, Wu X, Johnson J, Li Z, Liu J, Szabó PE, Lu Q, Xu GL, Song J, Pfeifer GP. 2016. Tet3 reads 5-carboxylcytosine through its CXXC domain and is a potential guardian against neurodegeneration. Cell Rep 14(3):493–505.
Jin SG, Xiong W, Wu X, Yang L, Pfeifer GP. 2015. The DNA methylation landscape of human melanoma. Genomics 106(6):322–330.
Jung M, Kadam S, Xiong W, Rauch TA, Jin SG, Pfeifer GP. 2015. MIRA-seq for DNA methylation analysis of CpG islands. Epigenomics 7(5):695–706.
Jung M, Pfeifer GP. 2015. Aging and DNA methylation. BMC Biol 13:7.
Jung M, Jin SG, Zhang X, Xiong W, Gogoshin G, Rodin AS, Pfeifer GP. 2015. Longitudinal epigenetic and gene expression profiles analyzed by three-component analysis reveal down-regulation of genes involved in protein translation in human aging. Nucleic Acids Res 43(15):e100.
Iqbal K, Tran DA, Li AX, Warden C, Bai AY, Singh P, Wu X, Pfeifer GP, Szabó PE. 2015. Deleterious effects of endocrine disruptors are corrected in the mammalian germline by epigenome reprogramming. Genome Biol 16:59.
Pfeifer GP. 2015. How the environment shapes cancer genomes. Current Opin Oncol 27(1):71–77.
Hahn MA, Li AX, Wu X, Pfeifer GP. 2015. Single base resolution analysis of 5-methylcytosine and 5-hydroxymethylcytosine by RRBS and TAB-RRBS. Methods Mol Biol 1238:273–287.
Mezzanotte J, Hill V, Schmidt ML, Shinawi T, Tommasi S, Krex D, Schackert G, Pfeifer GP, Latif F, Clark GJ. 2014. RASSF6 exhibits promoter hypermethylation in metastatic melanoma and inhibits invasion in melanoma cells. Epigenetics 9(11):1496–1503.
Hahn MA, Szabó PE, Pfeifer GP. 2014. 5-hydroxymethylcytosine: a stable or transient DNA modification? Genomics 104(5):314-323.
Dunwell TL, Pfeifer GP. 2014. Drosophila genomic methylation: new evidence and new questions. Epigenomics 6(5):459–461.
Fu L, Guerrero C, Zhong N, Amato N, Liu Y, Liu S, Cai Q, Jin SG, Ji D, Niedernhofer L, Pfeifer GP, Xu GL, Wang Y. 2014. Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J Am Chem Soc 136(33):11582–1585.
Hahn MA, Li AX, Wu X, Yang R, Drew DA, Rosenberg DW, Pfeifer GP. 2014. Loss of the polycomb mark from bivalent promoters leads to activation of numerous cancer-promoting genes in colorectal tumors. Cancer Res 74(13):3617–3629.
Dunwell TL, McGuffin LJ, Dunwell JM, Pfeifer GP. 2013. The mysterious presence of a 5-methylcytosine oxidase in the Drosophila genome: possible explanations. Cell Cycle 12(21):3357–3365.
Kalari S, Jung M, Kernstine KH, Takahashi T, Pfeifer GP. 2013. The DNA methylation landscape of small cell lung cancer suggests a differentiation defect of neuroendocrine cells. Oncogene 32(30):3559–3568.
Pfeifer GP, Kadam S, Jin SG. 2013. 5-hydroxymethylcytosine and its potential roles in development and cancer. Epigenetics Chromatin 6(1):10.
Hahn MA, Qiu R, Wu X, Li AX, Wang J, Zhang H, Jui J, Jin SG, Jiang Y, Pfeifer GP*, Lu Q*. 2013. Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Reports 3(2):291–300.
*Contributing authors, equal contributions
Lomniczi A, Loche A, Castellano JM, Ronnekleiv OK, Bosch M, Kaidar G, Knoll JG, Wright H, Pfeifer GP, Ojeda SR. 2013. Epigenetic control of female puberty. Nat Neurosci 16(3):281–289.
Zheng L, Dai H, Zhou M, Li X, Liu C, Guo Z, Wu Z, Wu J, Wang C, Zhong J, Huang Q, Garcia-Aguilar J, Pfeifer GP, Shen B. 2012. Polyploid cells rewire DNA damage response networks to overcome replication stress-induced barriers for tumor progression. Nat Commun 3:815.
Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, Krex D, Lu Q, Pfeifer GP. 2011. 5-hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res 71(24):7360–7365 (Priority Report).
Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, Iqbal K, Shi YG, Deng Z, Szabo PE, Pfeifer GP, Li J, Xu GL. 2011. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477(7366):606–610.
Jin SG, Wu X, Li AX, Pfeifer GP. 2011. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res 39(12):5015–5024.
Iqbal K, Jin SG, Pfeifer GP*, Szabó PE*. 2011. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci USA 108(9):3642–3647.
*Equally contributing authors
Guo C, Zhang X, Pfeifer GP. 2011. The tumor suppressor RASSF1A prevents dephosphorylation of the mammalian STE20-like kinases MST1 and MST2. J Biol Chem 286(8):6253–6261.
Pfeifer GP, Hainaut P. 2011. Next-generation sequencing: emerging lessons on the origins of human cancer. Curr Opin Oncol 23(1):62–68.
Kalari S, Pfeifer GP. 2010. Identification of driver and passenger DNA methylation in cancer by epigenomic analysis. Adv Genet 70:277–308.
Jin SG, Kadam S, Pfeifer GP. 2010. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res 38(11):e125.
Pfeifer GP, Dammann R, Tommasi S. 2010. RASSF proteins. Curr Biol 20(8):R344-R345.
Wu X, Rauch TA, Zhong X, Bennett WP, Latif F, Krex D, Pfeifer GP. 2010. CpG island hypermethylation in human astrocytomas. Cancer Res 70(7):2718–2727.
Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. 2009. A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci USA 106(3):671–678.
Hahn MA, Hahn T, Lee DH, Esworthy RS, Kim BW, Riggs AD, Chu FF, Pfeifer GP. 2008. Methylation of polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res 68(24):10280–10289.
Jin SG, Guo C, Pfeifer GP. 2008. GADD45A does not promote DNA demethylation. PLoS Genet 4(3):e1000013.
Rauch TA, Zhong X, Wu X, Wang M, Kernstine KH, Wang Z, Riggs AD, Pfeifer GP. 2008. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci USA 105(1):252–257.
Guo C, Tommasi S, Liu L, Yee JK, Dammann R, Pfeifer GP. 2007. The RASSF1A tumor suppressor protein is a component of a mammalian complex analogous to the Drosophila Hippo/Salvador/Lats tumor suppressor network. Curr Biol 17(8):700–705.
Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau S, Kernstine K, Riggs AD, Pfeifer GP. 2007. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci USA 104(13):5527–5532.
Rauch T, Li H, Wu X, Pfeifer GP. 2006. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res 66(16):7939–7947.
Liu L, Tommasi S, Lee DH, Dammann R, Pfeifer GP. 2003. Control of microtubule stability by the RASSF1A tumor suppressor. Oncogene 22(50):8125–8136.
Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. 2002. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene 21(48):7435–7451.
Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. 2000. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet 25(3):315–319.
Denissenko MF, Chen JX, Tang MS, Pfeifer GP. 1997. Cytosine methylation determines hotspots of DNA damage in the human P53 gene. Proc Natl Acad Sci USA 94(8):3893–3898.
Denissenko MF, Pao A, Tang MS, Pfeifer GP. 1996. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science 274(5286):430–432.
Tornaletti S, Pfeifer GP. 1994. Slow repair of pyrimidine dimers at p53 mutation hot spots in skin cancer. Science 263(5152):1436–1438.
Pfeifer GP, Steigerwald SD, Mueller PR, Wold B, Riggs AD. 1989. Genomic sequencing and methylation analysis by ligation mediated PCR. Science 246(4931):810–813.

Jennifer Brooks
Senior Administrative Assistant I, Department of Epigenetics
Wei Cui, Ph.D.
Research Scientist, Department of Epigenetics


Zhijun Huang, Ph.D.
Research Scientist, Department of Epigenetics

Seung-Gi Jin, Ph.D.
Senior Research Scientist, Department of Epigenetics

Jennifer Johnson, M.S.
Research Technician, Department of Epigenetics